





|
 |
 |
Synthetic Trinucleotide (codon & anticodon) phosphoramidites for Protein mutagenesis
Oligonucleotide-directed mutagenesis is probably the most popular approach for the preparation of proteins with variations at specific sites 1-5
In principle, the most flexible way to randomize oligonucleotides for generation of protein libraries would be the use of trimer phosphoramidites. Of the 64 possible combinations of codons, only 20 codons would be required to cover the 20 amino acids, although, in practice, several codons will likely be duplicated depending on the organism. Obviously, Trimer phosphoramidites offer an elegant solution that circumvents problems of codon bias, frame-shift mutations and stop-codon production, when compared to technique using mixtures of monomers to generate codon mixtures.
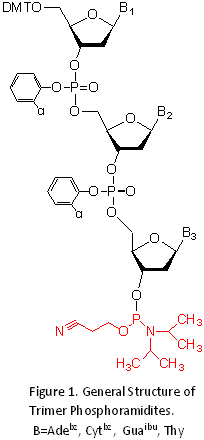
Several reports describing 6-8 the synthesis of trimer phosphoramidites have been published. General structure of
commercially available set of Trinucleotide phosphoramidites is shown in Figure 1. Once a mutagenic oligonucleotide library has been synthesized using Trimer Phosphoramidites, there are a variety of means introducing it into a plasmid for construction of a gene library.9. 9,10
"Codon" and "Anticodon" Trimers for protein mutagenesis. Why do we need "codon" and "anticodon" Trimers for randomization of Genes?
Nowadays full chemical synthesis of genes has become very common and widely commercially available. It is usually done through PCR extension of overlapping oligonucleotides, partially representing coding and noncoding strands of the required sequence.
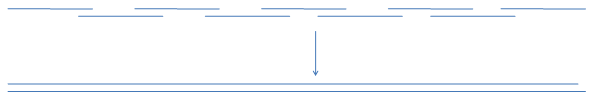
This is also a convenient way of assembling gene libraries where variable part (blue bar)
of the oligo library is positioned in the gap of the opposite strand to be filled with polymerase.
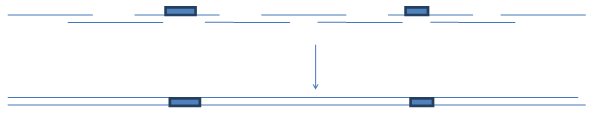
With earlier commercially available set of trimers (Metkinen Chemistry and Glen Research) it was only possible to introduce
variations through the coding strand. . And thus adjacent stretches of the sequence were sometimes difficult to randomize.
If the "anticodon" trimers, which can be used as part of complementary (noncoding) strand would be available, the following scheme could be implemented:
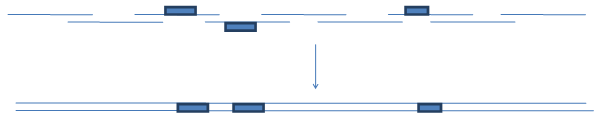
Similar scheme could be implemented in template-dependent overlapping PCR that will also allow an easy placement of variable regions in close proximity.

In order to develop an approach for Protein Mutagenesis, when oligonucleotides with randomizations could be designed as forward or reverse primers, the earlier available set of Trimer phosphoramidites
(Table 1) was not sufficient and had to be expanded.
Table 1. "Codon" Trimers
| 1 |
2 |
| Trimers for placing in coding (sense) strand |
Amino acid encoded |
| TAC |
Tyr |
| TCT |
Ser |
| GGT |
Gly |
| CGT |
Arg |
| GCG |
Ala |
| GAT |
Asp |
| AAC |
Asn |
| CAG |
Gln |
| GAA |
Glu |
| CAT |
His |
| ATC |
Ile |
| CTG |
Leu |
| AAA |
Lys |
| ATG |
Met |
| TTC |
Phe |
| CCA |
Pro |
| ACC |
Thr |
| TGG |
Trp |
| GTT |
Val |
| TGC |
Cys |
The new set incorporates optimal E.coli "codons" for all 20 aminoacids, when placed in the coding (sense) orientation (Table 1).
In addition, the new set also incorporates "anticodons" for placing in the non-coding (antisense) orientation.
These "anticodons", in turn will give rise to optimal "codons" upon replication. In other words, oligonucleotides with randomizations could be designed as forward or reverse primers.
Only 15 out of 20 "codon" Trimers could be employed as "anticodones" (Table 2, column 1, Trimers given in black),
that would be replicated to optimal E.coli codons (Table 2, column 2, Trimers given in bold italics)
and subsequently translated into the corresponding amino acids (Table 2, column 3).
Therefore 5 additional "anticodons" had to be added to the list (Table 2, column 1, Trimers given in red),
thus making it possible to use a set of 25 Trimers for randomization of both coding and noncoding DNA strands.
Tables 1 and 2 demonstrates these 25 "codons" and "anticodons" in more detail.
Table 2. "Antcodon" Trimers of MC
| 1 |
2 |
3 |
MC Trimer for introducing into
noncoding strand ("anticodon"
trimers) |
Trimer in coding strand resulting from
trimer, placed in the noncoding strand
upon replication |
Amino acid
encoded |
| GTA |
TAC |
Tyr |
| AGA
| TCT |
Ser |
| ACC |
GGT |
Gly |
| GCG |
CGC |
Arg |
| TGC |
GCA |
Ala |
| ATC |
GAT |
Asp |
| GTT |
AAC |
Asn |
| CTG |
CAG |
Gln |
| TTC |
GAA |
Glu |
| ATG |
CAT |
His |
| GAT |
ATC |
Ile |
| CAG |
CTG |
Leu |
| TTT |
AAA |
Lys |
| CAT |
ATG |
Met |
| GAA |
TTC |
Phe |
| TGG |
CCA |
Pro |
| GGT |
ACC |
Thr |
| CCA |
TGG |
Trp |
| AAC |
GTT |
Val |
| GCA |
TGC |
Cis |
Today all 25 Trimer (codon & anticodon) phosphoramidites are available from Metkinen Chemistry. In addition, Metkinen Chemistry is ready to perform a custom synthesis of randomized oligonucleotide libraries.
References
1. J. Sondek and D. Shortle, Proc Natl Acad Sci U S A, 1992, 89, 3581-3585.
2. P. Gaytan, J. Yanez, F. Sanchez, H. Mackie, and X. Soberon, Chem Biol, 1998, 5, 519-527.
3. P. Gaytan, J. Yanez, F. Sanchez, and X. Soberon, Nucleic Acids Res, 2001, 29, E9.
4. F.A. Fellouse, et al., J Mol Biol, 2007, 373, 924-40.
5. E-C. Brockmann, S. Akter, T. Savukoski, A. Lehmusvuori, J. Leivo, O. Saavalainen, A. Azhayev, T. Lövgren, J. Hellman, U. Lamminmäki, Protein Eng. Des.Sel. 2011, 1-10.
6. A.L. Kayushin, M.D. Korosteleva, A.I. Miroshnikov, W. Kosch, D. Zubov, and N. Piel, Nucleic Acids Research, 1996, 24, 3748-3755
7. T. Mauriala, S. Auriola, A. Azhayev, A. Kayushin, M. Korosteleva, and A. Miroshnikov, J.Pharmaceutical and Biomedical Analysis, J Pharm Biomed Anal, 2004, 34, 199-206.
8. A. Yagodkin, A. Azhayev, J. Roivainen, M. Antopolsky, A. Kayushin, A., M. Korosteleva, A. Miroshnikov, A., J. Randolph, H. Mackie, Nucleosides, Nucleotides and Nucleic Acids 2007, 473-497 and references cited therein.
9. J.Randolph, A.Yagodkin, M.Lamaitre, A.Azhayev, H.Mackie, Nucleic Acids Symposium Series 02/2008; DOI:10.1093/nass/nrn243
10. S.S. Sidhu, H.B. Lowman, B.C. Cunningham, and J.A. Wells, Methods Enzymol, 2000, 328, 333-63
|
 |
|

